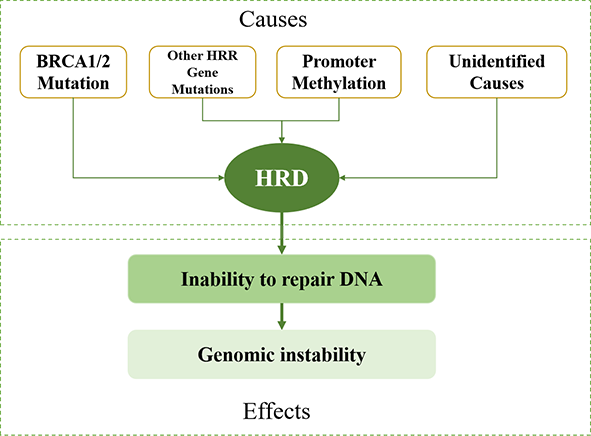
 How to determine HRD status?
How to determine HRD status?
by gene mutations, promoter methylation, or undetermined causes, the HR pathway stops working, leading to Homologous Recombination Deficiency (HRD).
for the assessment of HRD regardless of the specific cause.
The AmoyDx® HRD Focus Panel offers dependable and cost-effective solutions for evaluating PARP inhibitor (PARPi)-related biomarkers and advancing targeted treatment research in ovarian cancer. Leveraging proprietary HANDLE technology, the Genomic Scar Score (GSS) algorithm, and the AmoyDx NGS Data Analysis System (ANDAS), the HRD Focus assay can be seamlessly integrated into your laboratory workflow. Our methodologies enable streamlined end-to-end solutions, requiring less than 1 hour of hands-on operation, delivering high-performance assessment of genomic instability, and providing automated and secure data analysis.
Comprehensively Designed GSS Algorithm
The AmoyDx proprietary GSS algorithm is a machine learning-based model which assesses genomic instability by analyzing different types of copy number events across the genome.1

Longer PFS with PARPi Treatment for GSS-positive Group1
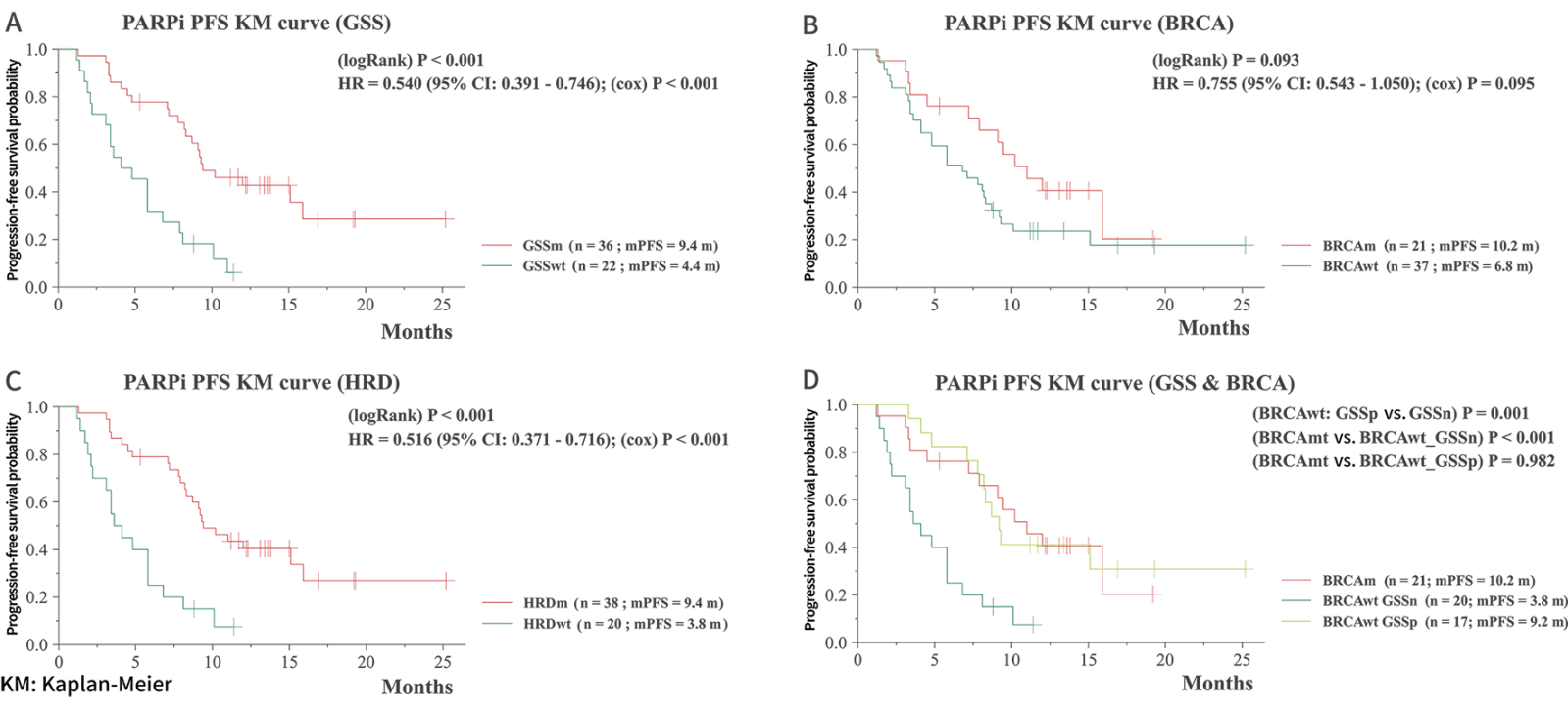 The study highlights the promising value of GSS in identifying patients
The study highlights the promising value of GSS in identifying patients who may respond favorably to PARPi treatment.

Specifications
Technology
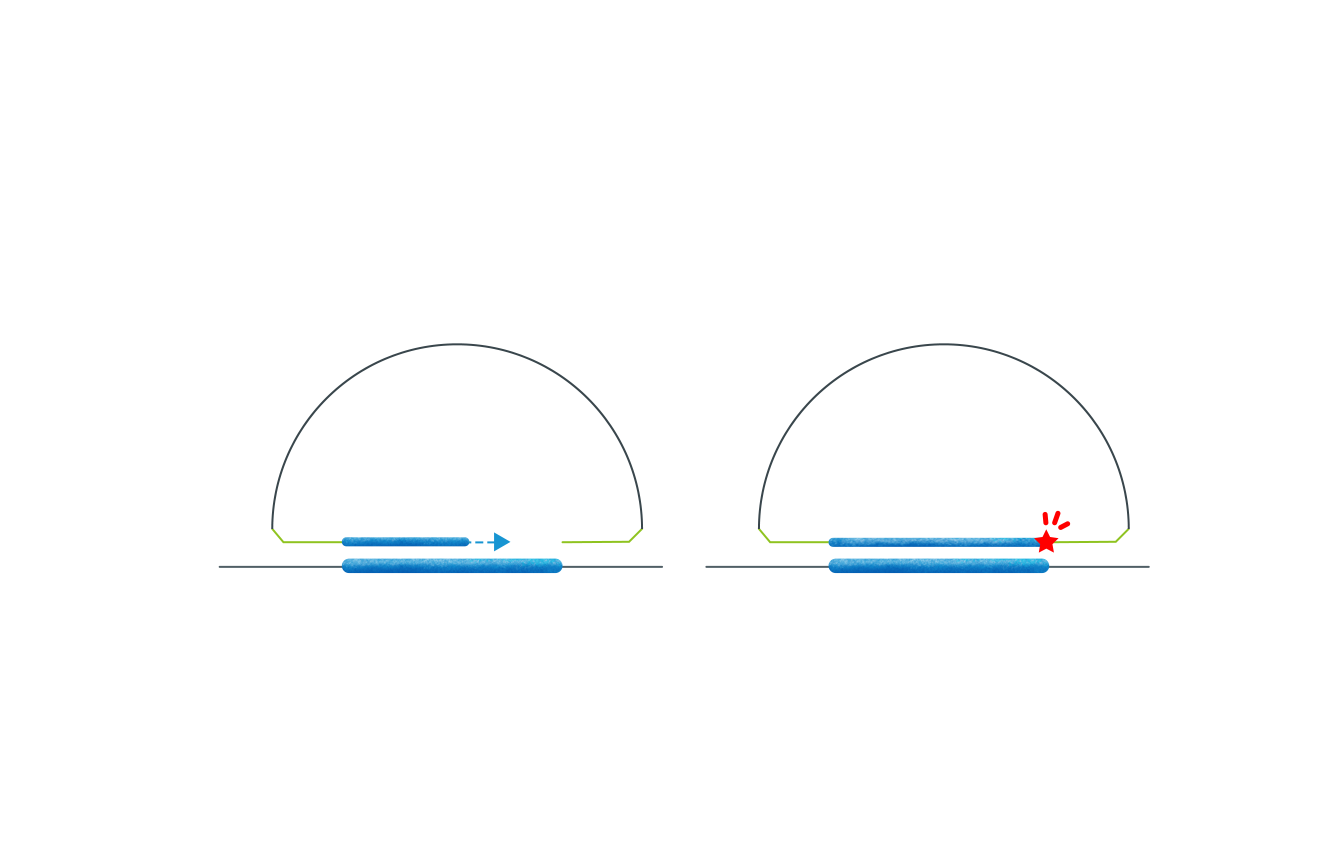
The AmoyDx HANDLE® technology is a proprietary amplicon-based method for NGS library preparation.
It offers a time-saving and cost-effective protocol that can be completed within 5 hours, requiring just about 1 hour of hands-on time.
During the library construction process, each individual DNA molecule is tagged with a unique molecular index (UMI) at both ends. This feature enables high sensitivity in variant detection by eliminating any library amplification and sequencing bias.
The probe consists of an extension arm and a ligation arm, both complementary to the target gene region.
Target-specific probes simultaneously hybridize to the DNA/cDNA fragment, forming a circular structure with the intended target captured between the probes. Subsequently, remaining linear probes, single-strand, and double-strand DNA are digested, leaving only the target circular DNA for PCR amplification to form the final library.
Publications


1. Fountzilas, E.; Papadopoulou, K.; Chatzikonstantinou, T.; Karakatsoulis, G.; Constantoulakis, P.; Tsantikidi, A.; Tsaousis, G.; Karageorgopoulou, S.; Koumarianou, A.; Mauri, D.; et al. Concordance between Three Homologous Recombination Deficiency (HRD) Assays in Patients with High-Grade Epithelial Ovarian Cancer. Cancers 2023, 15, 5525.
2. Kekeeva, T.; Andreeva, Y.; Tanas, A.; Kalinkin, A.; Khokhlova, S.; Tikhomirova, T.; Tyulyandina, A.; Popov, A.; Kuzmenko, M.; Volkonsky, M.; et al. HRD Testing of Ovarian Cancer in Routine Practice: What Are We Dealing With? Int. J. Mol. Sci. 2023, 24, 10497.
3. Magliacane, G.; Brunetto, E.; Calzavara, S.; Bergamini, A.; Pipitone, G.B.; Marra, G.; Redegalli, M.; Grassini, G.; Rabaiotti, E.; Taccagni, G.; et al. Locally Performed HRD Testing for Ovarian Cancer? Yes, We Can! Cancers 2023, 15, 43.
4. Doig, Kenneth D. et al. Homologous Recombination Repair Deficiency: An Overview for Pathologists. Modern Pathology, Volume 36, Issue 3, 100049
5. Fumagalli, C., Betella, I., Ranghiero, A., Guerini-Rocco, E., Bonaldo, G., Rappa, A., Vacirca, D., Colombo, N. and Barberis, M. 2022. In-house testing for homologous recombination repair deficiency (HRD) testing in ovarian carcinoma: a feasibility study comparing AmoyDx HRD Focus panel with Myriad myChoiceCDx assay. Pathologica - Journal of the Italian Society of Anatomic Pathology and Diagnostic Cytopathology. 114, 4 (Sep. 2022), 288-294.
6. Scheiter A, Hierl F, Winkel I, Keil F, Klier-Richter M, Coulouarn C, Lüke F, Kandulski A, Evert M, Dietmaier W, et al. Wnt/β-Catenin-Pathway Alterations and Homologous Recombination Deficiency in Cholangiocarcinoma Cell Lines and Clinical Samples: Towards Specific Vulnerabilities. Journal of Personalized Medicine. 2022; 12(8):1270.
7. Liu H, Zhang Z, Chen L, Pang J, Wu H and Liang Z (2022) Next-Generation Sequencing Reveals a Very Low Prevalence of Deleterious Mutations of Homologous Recombination Repair Genes and Homologous Recombination Deficiency in Ovarian Clear Cell Carcinoma. Front. Oncol. 11:798173.






